Identification of Bovine Mastitis Associated Pathogens by Multiplex PCR-Juniper Publishers
Journal of Dairy & Veterinary Sciences- Juniper Publishers

Introduction
Healthy udder of milking cows is one of the most
important factors of successful breeding from the health as well as
economic point of view. Mastitis is defined as an inflammation of the
parenchyma of mammary gland, which not only reduces milk yield but
alters milk composition [1]
also. It is one of the most problematic diseases and continues to have a
major economic impact on the dairy industry throughout the world [2].
It is a devastating disease haunting the dairy industry leading to
reservoir of infection for human beings and the economic losses due to
high morbidity, discarded milk, reduced milk production and increased
antimicrobial resistance of the organisms in animals treated with
antibiotics [3].
The genome sequences of many of the major mastitis causing pathogens
are now available and can be utilized to develop nucleic acid-based
testing methods. Nucleic acid testing by PCR is highly sensitive and
specific. The identification of the bacteria using PCR at the species
level is based on amplification of a target gene which is mostly rRNA
gene, highly conserved in bacteria. It is the only assay that allows
simultaneous screening for multiple pathogens that might be causing
mastitis. To improve the diagnosis a rapid, sensitive and specific
Multiplex polymerase chain reaction (mPCR) assay has been developed for
detection of more than one pathogen in single PCR reaction causing
mastitis at early stages. In multiplex PCR, multiple pairs of primers
specific for different DNA segments are included in the same reaction to
enable amplification of multiple target sequences in one assay [4]. This produces amplicons of variable sizes, allowing the identification of targeted bacterial pathogens in a milk sample.
Materials and Methods
Sampling and general microbiological analysis
A total of 50 milk samples from cattle each about
5-10 ml in amount were collected in sterilized test tubes. The milk
samples were collected from Veterinary Clinical Complex and dairy farm
in and around the Bikaner city, Rajasthan with clinical mastitis. The
samples were collected in the morning and were immediately taken
thereafter to the laboratory on ice for further processing.
Isolation and identification of bacteria
Culture characteristics: The milk samples were
subjected to aerobic cultivation. Each milk sample was streaked on
Nutrient agar, Blood agar and MacConkey (MCA) agar plates in primary,
secondary and tertiary fashion in order to obtain isolated colonies of
bacteria. These petri plates were incubated for 24hr at 37 °C. After
24hr incubation these isolated colonies were cultured on Mannitol salt
agar (MSA), Edward's medium agar, Eosine methylene blue agar (EMB),
Mac-conkeyand Cetrimide agar plates respectively for isolation of
i>Staphylococcus aureus, Streptococcus spp., Escherichia coli,
Klebsiella pneumoniae and Pseudomonas aeruginosa and incubated
for 24 hours at 37 °C. The growth was examined for the colonial
morphology and pigmentation, and different types of colonies were
sub-cultured on separate nutrient agar plates in order to obtain pure
cu.
Biochemical characteristics: Cultures were
subjected to primary biochemical tests viz. Catalase test, Oxidase
test,and secondary biochemical tests viz. Oxidation-Fermentation test,
Mannitol fermentation, Growth on Mac-Conkey agar, 6.5% Sodium Chloride
tolerance test, Aesculin Hydrolysis, Pigment production,
Gelatinliquefaction, IMViC tests, growth on TSI slants, Sugar
fermentation tests, Urease test, DNAse test, Arginine hydrolysis test,
Nitrate reduction test and haemolysis on blood agar etc.
Genotyping: 23S rRNA spacer region gene based identification was carried out for S. aureus, S. agalactiae, S. dysgalactiae, S. uberis and E. coli as per the method described by [5]. 16S rRNA gene based identification as per the method described by [6] for P aeruginosa. 16S-23S rDNA internal transcribed spacer (ITS) region based identification as per the method described by [7] for K. pneumonia (Table1).

Results
Out of the 50 processed clinical mastitic milk samples, 33 (66%) samples were found positive for the presence of bacteria viz. E. coli, S. aureus, P. aeruginosa, K. pneumoniae, S. agalactiae.
In remaining 17 (34%) samples of clinical mastitis, no aerobic bacteria
could be isolated. A total of 41 bacterial isolates could be cultured
and identified by biochemical and genotypic test which consist of
14(28%) isolates of E. coli, 12(24%) isolates of S. aureus, 9(18%) isolates of P aeruginosa, 5(10%) isolate of K. pneumonia and 1(2%) isolate of Streptococcus agalactiae.
All 14 isolates of E. coli on preliminary
biochemical characterization revealed characteristic IMViC pattern,
growth on TSI (Y/Y/-) and fermented different sugars. Preliminary
biochemical characterization of all 12 isolates of S. aureus
revealed characteristic growth pattern on mannitol salt agar, showed
haemolysis on blood agar, DNAse test and fermented different sugars. All
9 P aeruginosa isolates revealed standard biochemical
characterization as growth at 42 °C, pigment production, growth on
triple sugar iron (TSI) agar (R/R/-), citrate utilization, arginine
hydrolysis, gelatin liquefaction, haemolysis and nitrate reduction.
Negative reaction was observed for aesculin hydrolysis and urease tests.
All 5 isolates of K. pneumoniaeon preliminary biochemical
characterization revealed characteristic IMViC pattern, growth on TSI
(Y/Y/-) and fermented different sugars. One Streptococcus agalactiae
isolate showed dew drop like colonies on Edward's medium, 6.5% sodium
chloride medium, hydrolysedaesculin and fermented different sugars.
Molecular diagnosis of common bacterial pathogens directly from bovine clinical mastitic milk
All the 50 samples were subjected for DNA extraction
directly. A standard protocol was followed for the isolation of DNA from
milk sample which is simple and rapid method as per Nachimutu et al. [8] with slight modifications. The method was easy to perform and requires minimal sample manipulation.
All the established species primer pairs were used
for evaluation of 5 bacterial species of bacteria in multiplex PCR. The
results of multiplex PCR revealed simultaneous detection of E. coli and
P. aeruginosa (8%), E. coli and K. pneumoniae (10%), aeruginosa (2%). However S. aureus and S. agalactiae were not P. aeruginosa and K. pneumoniae (2%) and E. coli, K. pneumoni, P detected by multiplex PCR.
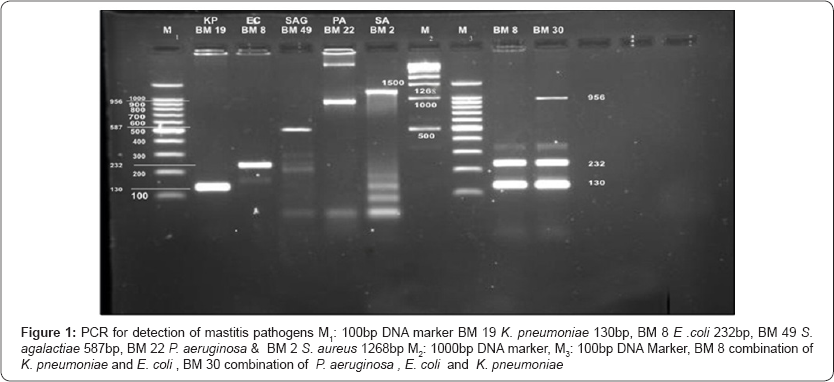
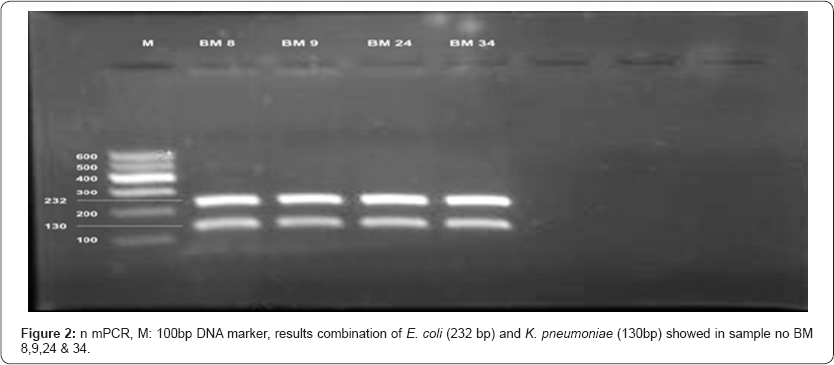
Comparative study of bacterial isolate, DNA with simplex PCR and DNA directly from milk with multiplex PCR
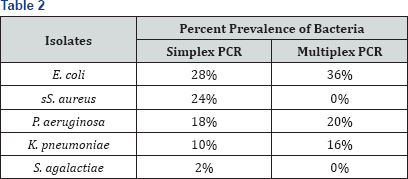
Discussion
Bovine mastitis is an important disease of cattle
which causes economic losses in the country by affecting the milk
quality and milk quantity. Mastitis is of multi-etiological in nature;
loss is due to the major bacterial pathogens, Staphylococcus aureus, Streptococcus spp, Escherichia coli, Klebsiella pneumoniae and Pseudomon asaeruginosa.
Diagnosis is more important in the case of mastitis. We can use
conventional method as culture and advanced method as molecular.
Compared to culture, molecular assays are less time consuming, takes
less than 24 hours to complete, while identification of bacteria to the
species levels by conventional microbiological and biochemical methods
requires more than 72 hours. Conventional procedures for the
identification of Bovine Mastitis pathogens are labor-intensive and most
of the commercial identification systems are not designed to identify
important mastitic pathogens [9,10]. In conventional method and molecular methods higher prevalence of E. coli found in the present study due to environment unhygienic condition which is somewhat similar with Bradely et al. [11]; Khaled et al. [12]; Sayed [13]
who have also reported higher incidence. The biochemical
characterization of isolates and fermentation of sugars are comparable
with studies of Kruthi [14] and Pachaury [15]. In the present investigation all E. coli
isolates were subjected to PCR amplification targeting 23S rRNA gene
using species specific primers. Species specific amplicon of 232bp are
similar with results of Riffon et al. [5] who found amplicon of this size. All 12 isolates of S. aureus
showed standard biochemical characterization and sugar fermentation
which similar with Upadhyay (2009) who carried out fermentation studies
with S. aureus obtained from milk of mastitic cattle and goat. Streptococcus spp. isolate biochemically confirmed and were subjected to PCR amplification targeting 23S rRNA spacer region by using S. agalactiae, S. dysgalactiaeand S. uberisspecies specific primers with 587bp, 403bp and 95bp. 23S rRNA spacer region based studies were conducted by Riffon et al. [5]
for genotypic confirmation of Streptococcus agalactiae isolated from
bovine mastitic milk and they observed 587 bp PCR product. All 8
isolates which had been biochemically identified as P aeruginosa
were subjected to genotyping using species specific primers reported
earlier (Spilkeret al., 2004). In the present investigation all 5
isolates had been biochemically identified as K. pneumonia and
were subjected to PCR amplification targeting 16S-23S rDNA internal
transcribed spacer (ITS) region based specific primers [7]. Our results are completely in agreement with [7].
As the present study is focused on development of a
rapid and early detection method, a multiplex PCR was devised to allow
the simultaneous detection of multiple pathogens of mastitis in a single
reaction, which requires smaller amounts of reagents and even lesser
time than simplex PCR [5,16].
This multiplex PCR method appeared as a useful tool to resolve
bacterial aetiology of mastitis milk samples simultaneously and at the
earliest [12,17,18].
Multiplexing is alternative method however, the
compatibility of the primer pairs is important for the reaction to work
efficiently with high sensitivity of detection. All the established
species primer pairs were used for evaluation of 5 bacterial species in
multiplex PCR. The results of multiplex PCR revealed simultaneous
detection of E. coli and P. aeruginosa (8%), E. coli and K. pneumoniae (10%), P aeruginosa and K. pneumoniae (2%) and E. coli, K. pneumoniae and P aeruginosa (2%) which is compare with Pradhan 2011 who used multiplex PCR as routine detection of S. aureus, Streptococcus spp. and E. coli in milk samples and However, S. aureus and S. agalactiae were not detected by multiplex PCR but various previous studies as done by Shome et al. [19]
were able to detect 10 bacterial strains simultaneously by multiplex
PCR. This disparity in results of simplex and multiplex PCR might be due
to incompatibility of primers or differences in annealing temperatures.
The multiplex PCR assay developed in this study provides a convenient
means of accurately identifying important bacterial pathogens of
mastitis simultaneously.
In cultural isolation, colonies are selected based on
phenotypic characteristics like size, color, shape etc. There can be
possibility of missing out colonies that are having similar phenotype to
the predominant ones but very few in numbers. Similarly, possible
reasons for no growth in milk samples can include low concentration of
bacteria in the milk samples, pathogens not growing in standard culture
media, or presence of substances in the milk decreasing the viability of
bacteria in culture [20].
When used for the detection of pathogens in subclinical mastitis milk
samples, mPCR was found to be more efficacious than microbiological
studies. The mPCR successfully detected all bacteria, as isolated in
culture method. Besides, mPCR detected organisms in samples found
culture-negative. Culture-negative milk samples in conventional
bacteriology represent a large proportion of positive samples in
molecular methods [20].
To know more about journal of veterinary science impact factor: https://juniperpublishers.com/jdvs/index.php
To know more about Open Access Publishers: Juniper Publishers


Comments
Post a Comment