Enterobacteriaceae in Some Imported Fish-Juniper Publishers
Journal of Dairy & Veterinary Sciences- Juniper Publishers

Introduction
Unlike meat and poultry, fish are more liable to
contamination with pathogenic bacteria from human reservoir which may
contaminate the water depending on the fishing and also may be further
contaminated during handling, processing and packaging. While the muscle
flesh of fish, which is the main edible part is normally sterile but
microorganisms can penetrate from the skin and the gut to the flesh, the
penetration and contamination increase in case of fish caught from
polluted area where there are high densities of bacteria. Singh &
Kulshrestha [1] isolated 17 strain of E.coli from fresh and marine fish, shrimp and mollusk fish which positive for enterotoxogenicity. Edris [2]
Samples collected from Cairo and Giza supermarkets included (Mackerel,
Sardine, Sardinella, fish burger and breaded shrimp). Such quality
control was evaluated through organoleptical, microbiological and
chemical examinations of 30 random samples of each fish and fish
products. The present investigation proved that the examined random
samples of the imported fish and fish products were quite safe for human
E. coli and Salmonella were not isolated from all the examined samples. Ahmed [3]
62 samples of chilled fish fillets, 50 samples of iced peeled shrimp
and 15 samples each of frozen imported and local peeled shrimp samples
were collected from Cairo and Giza markets. Examined samples for
isolation and identification of specific pathogens Vibrio spp., E. coli, Listeria spp., A. hydrophila, S. aurerus and Y. enterocolitica,
Salmonella and Shigella. About 82.3% of filets samples were accepted
according to the ESS (3494/2005), while 72% of peeled shrimp samples
were accepted according to ESS (516/1993), whereas, 100 and 93% of
frozen imported and local shrimp samples were accepted. E. coli were isolated from fillets and peeled shrimp samples. Khadega [4] isolated Salmonella from10% and 16% from Mullus and Basa while she failed to isolate Salmonella from Barboni. Elhadi [5] examined 35 samples and found 11 samples contaminated with Salmonella (31.4%). Morris et al. [6]
examined samples of fish immediately after catching where they failed
to isolate Salmonella. But after they arrived to the plants, they could
isolate this organism. Based on Morris's research imported fish
contaminated during transportation, packaging and handling process.
Onyango [7] among 120 imported Basa, 63 (52.5%) were infected with Enterobacteriaceae. Out of these, 25 (39.7%) were Shigella spp, 9 (14.3%) Salmonella typhimurium, 7 (11.1%) S. typhi, 4 (6.3%) S. enteritidis, 16 (25.4%) Escherichia coli, 1 (1.6%) Proteus spp. and Enterobacter aerogenes, respectively. Ten fish collected from open-air markets yielded E. coli (50%), S. typhimurium (20%),
S. paratyphi (10%) and S. typhi (20%). El-Leboudi [8] examined 15 imported fish samples, the author isolated 2 of Yersinia enterocolitica strains (13.3%) from imported fish. Harydi [9]
a total of 675 imported frozen fish samples from different origins were
collected on arrival to Sokhna port, Suez city, Egypt. The prevalence
of Salmonella spp. were found to be (1.6%) in whole shrimp,
(3.7%) in peeled shrimp, (2.0) in whole fish fillets, and (1.67%) in
sepia, respectively. Meanwhile in calamari; Salmonella prevalence was
found to be 4.0%. Alzainy [10] examined 60 samples of frozen imported fish and live fish, 65% of samples were found to be positive for Aeromonas hydrophila
isolation 76.6% were in life fish samples and 53.3% in frozen fish
94.87% exhibited α and β hemolysis, 100% of life fish isolates show p
hemolysis while frozen fish isolates show 85.7% β hemolysis and 14.3% α
hemolysis, 97.43% of isolates show cytotoxic effect on Vero cells the
highest frequency occur in the isolates of life fish group 60.50%. Farag
[11]
Sixty two samples of chilled fish fillets, 50 samples of iced peeled
shrimp and 15 samples each of frozen imported and local peeled shrimp
samples were collected from Cairo and Giza markets bacteria counts for Aeromonas spp and vibrio spp. Were positive. Sharma [12]
the research has shown that imported fish is most frequently and most
extensively contaminated with bacteria from the Aeromonas genus
(positive samples ranging from 37% to 93%). El Noby [13] Sixty samples (20 each of Tilapia sp., Mugil cephalus
and frozen mackerel fish samples) were randomly collected from
different local shops of fish sailing and fish retailers of different
sanitation levels at Zagazig city. Collected samples were examined
bacteriologically for determination of the incidence as well as the
count of psychrotrohic microorganisms using the rapid method (25 pOCI 24
hours) following by pour plate technique. The obtained results revealed
that. The mean count of Aeromonas was 5.7x10)±3.0x102, 1.0x105±4.9x104
and 3.3x105±2.7x104cfu/g of examined Tilapia nilotica, Mugil cephalus and frozen Mackerel samples, respectively.
Materials and Methods
Materials
Collection of samples: A total of 200 random samples of imported fishes represented by Bangasius hypophanmus (Basa), mullus surmuletus (Barboni), Saurida undosquamis (Mackerel) and Sparus aurata
(Denise) (50 of each) were collected from different fish markets in
Alexandria city. Each sample (250g) was kept in a separate plastic bag
and transferred directly with a minimum of delay to the laboratory of
food hygiene department, Faculty of Veterinary Medicine, Alexandria
University in an insulating refrigerated container under complete
aseptic condition to avoid any changes in the quality of the sample.
Samples were examined bacteriologically immediately after arrival to the
laboratory for isolation and identification of Salmonella, Escherichia coli, Shigella, Vibrio parahaemolyticus, Yersinia enterocolitica and Aeromonas.
Culture media: EMB medium (HIMEDIA), XLD agar
(Acumedia), MacConkey, Agar (BBL), Nutrient Agar (Difco), Semisolid agar
(Difco), Thiosulfate-citrate-bile salt-sucrose agar (TCBS Agar) (BBL),
M-Aeromonas Selective Agar Base (Havelaar) (Himedia) with ampicillin
vial, Cefsulodin-irgasan-novobiocin (CIN) agar (Oxoid), Trypticase Soy
Broth, Bile-Oxalate-Sorbose broth and MacConkey's broth.
Methods
25 grams of each fish fillet sample were aseptically
transferred into sterile blender flask containing 225ml of sterile
peptone water 0.1% and homogenized at 1400rpm for 2-5 minutes to provide
a homogenate 1/10 dilution and then allowed to stand for about 6
minutes at room temperature. The contents of the flask were thoroughly
mixed by shaking and one ml of the homogenate was transferred with
sterile pipette to another tube containing 9 ml of sterile peptone
water, from which tenth fold serial dilutions were prepared up to 10-6
(APHA, 1985) [14].
1.Isolation and identification of Salmonellae: According to (Rappaport & Harvy and Price) [15,16], (Collins & Cruickshank) [17,18], (Simmon) [19], (Kovacs) [20], (Ljutov) [21], (MacFaddin) [22], (Hugh & Leifson) [23], (Edwards & Ewing) [24], (Kauffman) [25] for serological examination, and (ICMSF, 1996) [26] for preenrichment.
2. Isolation and identification of Escherichia coli:
According to (Rappaport & Harvy) [15,16], (Collins & Cruickshank) [17,18], (Simmon) [19], (Kovacs) [20], (Ljutov) [21], (MacFaddin) [22], (Hugh & Leifson) [23], and (Edwards & Ewing) [24].
3. Isolation and identification of Shigella: According to (Rappaport & Harvy) [14,15], (Collins & Cruickshank) [17,18], (Simmon) [19], (Kovacs) [20], (Ljutov) [21], (MacFaddin) (Hugh & Leifson) [23], and (Edwards & Ewing) [24].
4. Isolation and identification of Yersinia: According to (Schiemann, 1983)[27] for Pre-enrichment, Krieg & Holt [28] and (MacConkey) [29] for selective enrichment and plating.
5. Isolation and identification of Vibrio parahaemolyticus: According to (APHA) [30].
6. Isolation and identification of Aeromonas: According to (Havelaar) [31].
7. The obtained results were statistically evaluated according to the guidelines recommended by Feldman et al. [32].
Discussion
Literature extended over many years pointed out that
fish and its products are liable to contamination with various kinds of
micro-organisms from different sources. Such contamination may render
the fish unsafe to the consumers or impair its utility, especially in
undeveloped countries, where the hygienic measures are still underway.
Many efforts were done to keep the fish free from pathogens of public
health hazard.
Isolation of enteropathogenic E. coli

It was evident from the results recorded in (Table 1), enteropathogenic E. coli was isolated from (21) 42%, (18) 36%, (19) 38% and (13) 26% of the examined samples of 5 Bangasius hypophanmus (Basa), mullus surmuletus (Barboni), 5aurida undosquamis (Mackerel) and 5parus aurata (Denise), respectively. This result was not compatible with the Egyptian standards (E.S889/2009) [33] of frozen fish, part 1 whole fish that stated that fish must be free from E. coli. The current result of isolation of E. coli from the examined samples of imported fish was higher than those obtained by Singh & Kulshrestha [1]
who could isolate 17 strains from all examined samples. Also, our
results for Mackerel is contradict the result obtained by Edris [2] who stated that E.coli was quite safe for human consumption according to his results. As well as Ahmed [3] who examined 15 samples of imported fish and stated that it is accepted for (E.S889/2009) [32]. Serotyping of enteropathogenic E.coli isolated from the examined samples of imported fish was declared in Table 2.
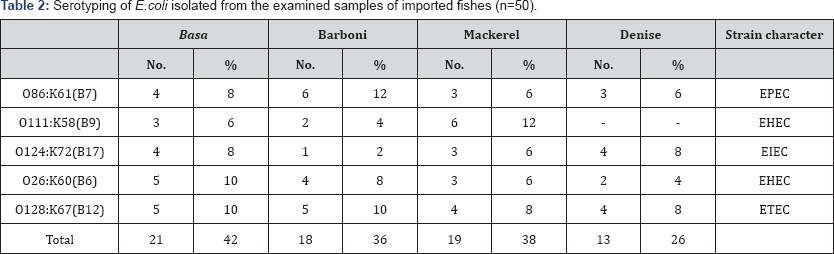
Table 2 Serotyping of E.coli isolated from the examined samples of imported fishes stated 5 strains of E. coli isolated from Basa,
Barboni and Mackerel, as O86:K61 (B7), O111:K58 (B9), O124:K72 (B17),
O26:K60(B6), and O128:K67(B12). Furthermore, 4 strains were
serologically isolated from Denise as O86:K61 (B7), O124:K72 (B17),
O26:K60 (B6), and O128:K67 (B12). The result in (Table 2)
shows the percentage of the incidence for each strain as follow, for
O86:K61(B7) it was (4) 8%, (6) 12%, (3) 6%, and (3) 6% of the examined
samples of S Bangasius hypophanmus (Basa), Mullus surmuletus (Barboni), Saurida undosquamis (Mackerel) and Sparus aurata (Denise), respectively. Secondly, incidence of O111:K58(B9), was (3) 6,(2) 4, (6) 12, and zero in the examined samples of S Bangasius hypophanmus (Basa), Mullus surmuletus (Barboni), Saurida undosquamis (Mackerel) and Sparus aurata (Denise), respectively Thirdly, incidence of O124:K72 (B17) was (4) 8, (1) 2, (3) 6, and (4) 8 in the examined samples of S Bangasius hypophanmus (Basa), Mullus surmuletus (Barboni), Saurida undosquamis (Mackerel) and Sparus aurata
(Denise), respectively. As well as, the incidence of O26:K60 (B6), was
(5) 10, (4) 8, (3) 6, and (2) 4 in the examined samples of S Bangasius hypophanmus (Basa), Mullus surmuletus (Barboni), Saurida undosquamis (Mackerel) and Sparus aurata (Denise), respectively Finally, incidence of O128:K67 (B12) was (5) 10, (5) 10, (4) 8, and (4) 8 in the examined samples of S Bangasius hypophanmus (Basa), Mullus surmuletus (Barboni), Saurida undosquamis (Mackerel) and Sparus aurata (Denise), respectively. Nearly similar results were reported by Donenberg & Kaper [33,34].
Salmonella
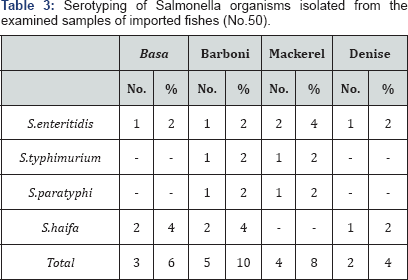
The recorded results in Table 3
stated incidence of Salmonella which was (3) 6%, (5) 10%, (4) 8% and
(2) 4% of the examined samples of Basa, Barboni, Mackerel and Denise,
respectively. This result was not compatible with the Egyptian standards
(E.S889/2009) [33] of frozen fish that stated that fish must be free from Salmonella in 25g. These results were in harmony with that of Khadega [4]
who could isolate Salmonella from10% and 16% from Mullus and Basa while
she failed to isolate Salmonella from Barboni. Also, Salmonella
organisms were previously isolated from imported fish by Stevens et al. [35], Baquar et al. [36] and Dalsgaarg [37]. Elhadi [5]
his result higher than our result in Mackerel, he examined 35 samples
and found 11 samples contaminated with Salmonella (31.4%), whereas, we
found only 4 out of 50 Mackerel samples (8%). We found Salmonella in 4
samples 8% of Mackerel, while Edris [2] couldn't find Salmonella in (30) examined Mackerel samples. As well as Ahmed [3] who stated that 15 samples of imported fish were free from Salmonella. It is important to mention that Morris et al. [6]
examined samples of fish immediately after catching where they failed
to isolate Salmonella. But after they arrived to the plants, they could
isolate this organism. Based on Morris's research imported fish
contaminated during transportation, packaging and handling process.
Accordingly, the presence of Salmonella as
enteropathogens in imported fish may reflect the unsatisfactory hygienic
conditions during handling, packaging and marketing of the fish.
Serological identification of the obtained Salmonella isolates was
tabulated in Table 2. It reflected that Sal. enteritidis (1) 2% and Sal. haifa (2) 4% were serologically identified from the examined samples of Basa. On the other hand, Barboni was contaminated by 4 different strains of Salmonella as follow, Sal. enteritidis (1) 2%, S. typhimurium (1) 2%, S. paratyphi (1) 2% and Sal. haifa (2) 4%. In addition, Denise was contaminated by 3 different strains of Salmonella as follow, Sal. enteritidis (2) 4%, S. typhimurium (1) 2%, and S.paratyphi (1)2%. Lastly, Serotyping of the isolated Salmonella indicated that Sal. enteritidis (1) 2% and Sal. haifa(1) 2% were serologically identified from the examined samples of Mackerel.
Shigella
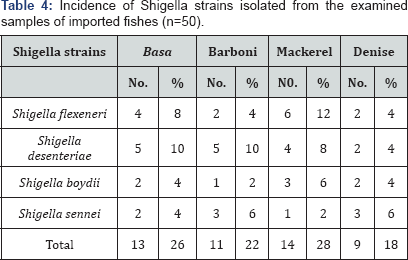
The illustrated data in Table 4
showed that the incidence of Shigella species in the examined imported
fish samples was (13) 26%, (11) 22%, (14) 28% and (9) 18% of the
examined samples of Basa, Barboni, Mackerel and Denise, respectively In
addition, the serotyping identification of the recovered isolates Table 3
revealed the examined samples of Basa were contaminated by Shigella
flexeneri (4) 8%, Shigella desenteriae (5) 10%, Shigella boydii (2) 4%
and Shigella sennei (2) 4%. Also, Barboni samples were contaminated by
Shigella flexeneri (2) 4%, Shigella desenteriae (5) 10%, Shigella boydii
(1) 2% and Shigella sennei (3) 6%. Furthermore, Barboni samples were
contaminated by Shigella flexeneri (2) 4%, Shigella desenteriae(5) 10%,
Shigella boydii (1) 2% and Shigella sennei (3) 6%.Furthermore, Mackerel
samples were contaminated by Shigella flexeneri (6) 12%, Shigella
desenteriae (4) 8%, Shigella boydii (3) 6% and Shigella sennei (1) 2%.
Finally, Denise samples were contaminated by Shigella flexeneri (2) 4%,
Shigella desenteriae (2) 4%, Shigella boydii (2) 4% and Shigella sennei
(2) 6%. According to (E.S889/2009) [38]
which assumed that frozen fish must be free from Shigella in 25g.
Consequence, these examined fish is unaccepted, on the other hand Ahmed [3]
who stated that 15 samples of imported fish were free from Shigella and
accepted. Imported Basa fish was contaminated by Shigella 26%, Onyango [7] among 120 imported Basa out of these, 25 (39.7%) were contaminated by Shigella spp.
Yersinia
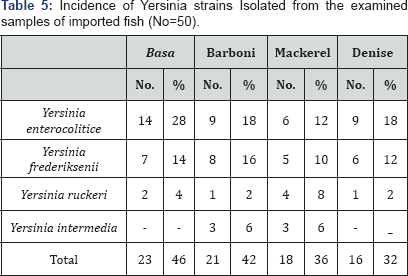
The illustrated data in Table 3 described Incidence of Yarsinia which was (23) 46%, (21) 42%, (18) 36% and (16) 32% of the examined samples of Basa, Barboni, Mackerel and Denise, respectively. In addition, the serotyping identification of the recovered isolates (Table 5) revealed the incidence of different strains of Yarsinia as follow, the examined samples of Basa contaminated by were Yarsinia enterocolitice (14) 28%, Yarsinia frederiksenii (7) 14%, Yarsinia ruckeri (2) 4% and Yarsinia intermedia 0%. Also, Barboni samples contaminated were Yarsinia enterocolitice (9) 18%, Yarsinia frederiksenii (8) 16%, Yarsinia ruckeri (1) 2% and Yarsinia intermedia (3) 6%. Furthermore, Mackerel samples were Yarsinia enterocolitice (6) 12%, Yarsinia frederiksenii (5) 10%, Yarsinia ruckeri (4) 8% and Yarsinia intermedia (3) 6%. Finally, Denise samples were Yarsinia enterocolitice (9) 18%, Yarsinia frederiksenii (6) 12%, and Yarsinia ruckeri (1) 2%. El-Leboudi [8] examined 15 imported fish samples, the author isolated 2 of Yersinia enterocolitica strains (13.3%) from imported fish. It is of great concern to record that Y. enterocolitica
was one of human pathogens that can grow at refrigeration temperature
and its presence in food constitutes a public health hazard. In this
respect, Y. enterocolitica has been implicated in several outbreaks of food illness during the past 20 years in numerous countries all over the world [37].
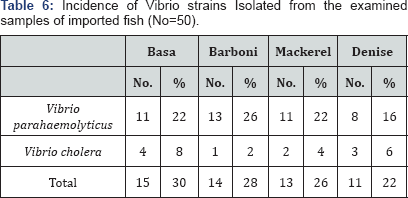
Vibrio spp
V cholera has long been known to be
responsible for the life threatening secretary diarrhea termed as
Asiatic cholera or epidemic cholera Ryan & Ray [38]. The illustrated data in Table 3 explained the incidence of Vibrio spp was (15) 30%, (14) 28%, (13) 26% and (11) 22% ofthe examined samples of Basa, Barboni, Mackerel and Denise, respectively. In addition, the serological identification of the recovered isolates (Table 4) showed the incidence of different strains of Vibrio as follow, the examined samples of Basa contaminated by Vibrio parahaemolyticus were (11) 22% and (4) 8% for Vibrio cholera. Also, Barboni samples contaminated were Vibrio parahaemolyticus were (13) 26% and (1) 2% for Vibrio cholera. Furthermore, Mackerel samples were Vibrio parahaemolyticus were (11) 22% and (2) 4% for Vibrio cholera. Finally, Denise samples were Vibrio parahaemolyticus were (8) 16% and (3) 6% for Vibrio cholera. Egyptian standards (E.S889/2009) [32] of frozen fish stated that fish must be free from V parahaemolyticus. V parahaemolyticus was previously isolated from imported fish by Abdelnoor & Roumani [39], Binta et al. [ 40] and Harydi [9].
Aeromonas hydrophilia
The illustrated data in Table 6 incidence of Aeromonas hydrophilia was (8) 16%, (11) 22%, (8) 16% and (6) 12% of the examined samples of Basa, Barboni, Mackerel and Denise, respectively. Also, Aeromonas hydrophilia founded in frozen imported fish samples by El-Noby [13], Vila et al. & Farag [11], Sharma [12] and Alzainy [10].
Finally, it is important to mention that the results
show that the higher bacterial count (Salmonella) was found in Barboni
and gently went down in Mackerel, Basa and Denise, respectively.
The higher bacterial count (E. coli) was found in Basa and gently went
down in Mackerel, Barboni and Denise, respectively. Moreover, results
show that the higher bacterial count (Shigella) was found in Mackerel
and gently went down in, Basa, Barboni and Denise, respectively [41,42].
Also, results show that the higher bacterial count (Yarisina) was found
in Basa and gently went down in Barboni, Mackerel and Denise,
respectively. The higher bacterial count (Vibro spp) was found in Basa
and gently went down in Barboni, Mackerel and Denise, respectively.
Finally, the higher bacterial count (Aeromonas hydrphilie) was found in
Barboni and gently went down in, (Basa and Mackerel) and Denise,
respectively. To sum up, it is obvious that Denise spp. was the lowest
contaminated imported fish for all kind of examined Enteropathogens. On
the opposite, Basa was the most contaminated spp. of imported
fish, except contamination by Salmonella it is clear that Barboni and
mackerel higher than Basa. Accordingly, the consumption of such
contaminated imported fish may, at times, induce public health hazard.
The obtained results in the current work, clarified that imported fish
possess a higher number of enteric pathogens with significant public
health risk. These results may be attributed to unsanitary conditions,
cross contamination, fecal pollution and bad personal hygiene conditions
during handling, storage, distribution and selling.
To know more about journal of veterinary science impact factor: https://juniperpublishers.com/jdvs/index.php
To know more about Open Access Publishers: Juniper Publishers


Comments
Post a Comment